Science
Related: About this forumMeasuring Foreign Chemical Exposure of Human Flesh: Mass Spectrometry Software for the "Exposome."
The paper I'll briefly discuss in this post is this one: Scannotation: A Suspect Screening Tool for the Rapid Pre-Annotation of the Human LC-HRMS-Based Chemical Exposome, Jade Chaker, Erwann Gilles, Christine Monfort, Cécile Chevrier, Sarah Lennon, and Arthur David, Environmental Science & Technology 2023 57 (48), 19253-19262.
Mass spectrometry is the very important technique of placing molecules in the gas phase - most often by spraying them in solution and evaporating off the solvents - and then adding energy to break them into ionic fragments (MS1) that can be electromagnetically stored, and fragmented again, (MS2) or even multiple times (MSn). The way molecules fragment, and the mass of the fragments, and now, even their shape (IMS, ion mobility spectrometry) is characteristic of their structure, and thus molecules can be, in practically every case with few exceptions, uniquely identified.
The technology of mass spectrometry has reached a level where one can almost determine the presence, and often the concentration, of molecules, both natural and foreign, at the femtogram and even the attogram levels but the data set sizes in doing so are massive, far too massive in fact, for a human being to apprehend in any meaningfully reasonable amount of time. There is even talk these days of identifying single molecules in this way.
As I recently noted in this space, referring to another paper in this issue of Environmental Science & Technology , the number of chemicals now in circulation for use for technological applications are huge: Conflicts of Interest in the Assessment of Chemicals, Waste, and Pollution
A quotation from the paper discussed in that post:
The paper cited at the outset of this post considers the question of how many, and what types, of industrial and consumer foreign molecules - generally falling under the rubric of "xenobiotics" - show up in human flesh by developing a software tool to sort through the complex data sets generated by mass spectrometry. This concept is defined as the "exposome," a concept defined in 2005 by the editor of a scientific journal, Cancer Epidemiology, Biomarkers & Prevention.
From the introduction to the paper mentioned at the outset:
Annotation of complex HRMS data can be performed using NTS, which relies on the structure elucidation of features, prioritized as differential between two (or more) groups, or SS, which relies on the annotation of features prioritized for their similarity to compounds listed in a suspect library. This second methodology is particularly promising, in part because it has a strong potential for automation and allows for a very rapid prioritization of signals of interest. Furthermore, there is no restriction regarding the number of suspects that can be included as well as the forms they are searched as (i.e., parent or metabolite, adduct). The comparison of experimental features and suspects on characteristic properties such as their mass/charge ratio or their MS2 fragmentation profile can be automatically performed before being manually validated. Even though some bioinformatics solutions already exist to carry out MS2-based spectral library searches, allowing the automatized comparison of reference and experimental MS2 spectra (e.g., xMSannotator, msPurity, MZmine2, MS-DIAL4, patRoon, CAMERA), (8−13) the number of software tools is still limited and not necessarily suited for exposomics applications, (14) which present specific challenges detailed hereafter.
One of the main challenges of using biological matrices to characterize the human internal chemical exposome is the wide dynamic range of concentrations for compounds present in the samples. Compounds of interest in an exposomics context are often present at much lower levels (i.e., tens of pg/mL (15)) than many endogenous metabolites (i.e., up to a few mg/mL). These low-abundant xenobiotics do not systematically trigger MS2 acquisition. (16) This strongly limits the annotation’s confidence level according to Schymanski’s scale, which is the current reference. (17) Despite this fact, other factors accessible through MS1 data, such as retention time (Rt), distinctive isotope profiles based on halogen contents (often present in exogenous compounds such as pesticides), or detection of other phase I/II metabolites, could already provide reliable indications of the annotation’s plausibility...
The authors use a lower level high resolution mass spectrometer, (HRMS) still a very powerful instrument but one that fits nicely on a benchtop to collect and annotate some data from human subjects.
A few graphics from the paper demonstrate the power of this software tool, which apparently can be downloaded for free by mass spectrometry labs:
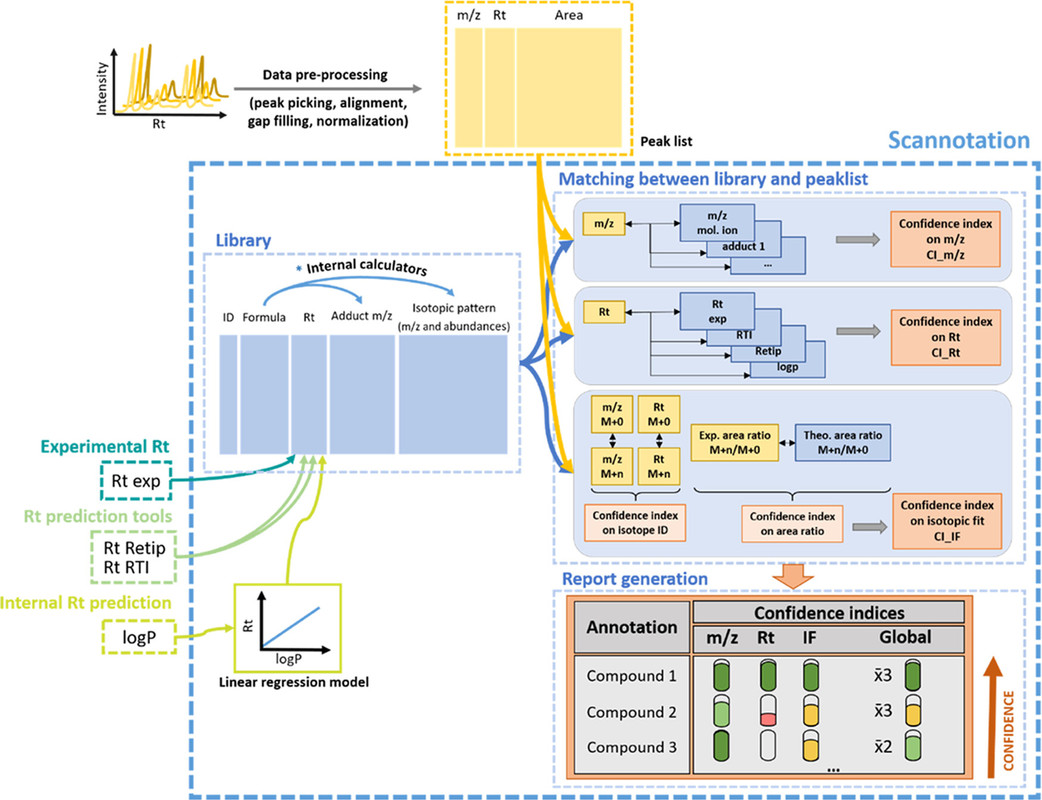
The caption:
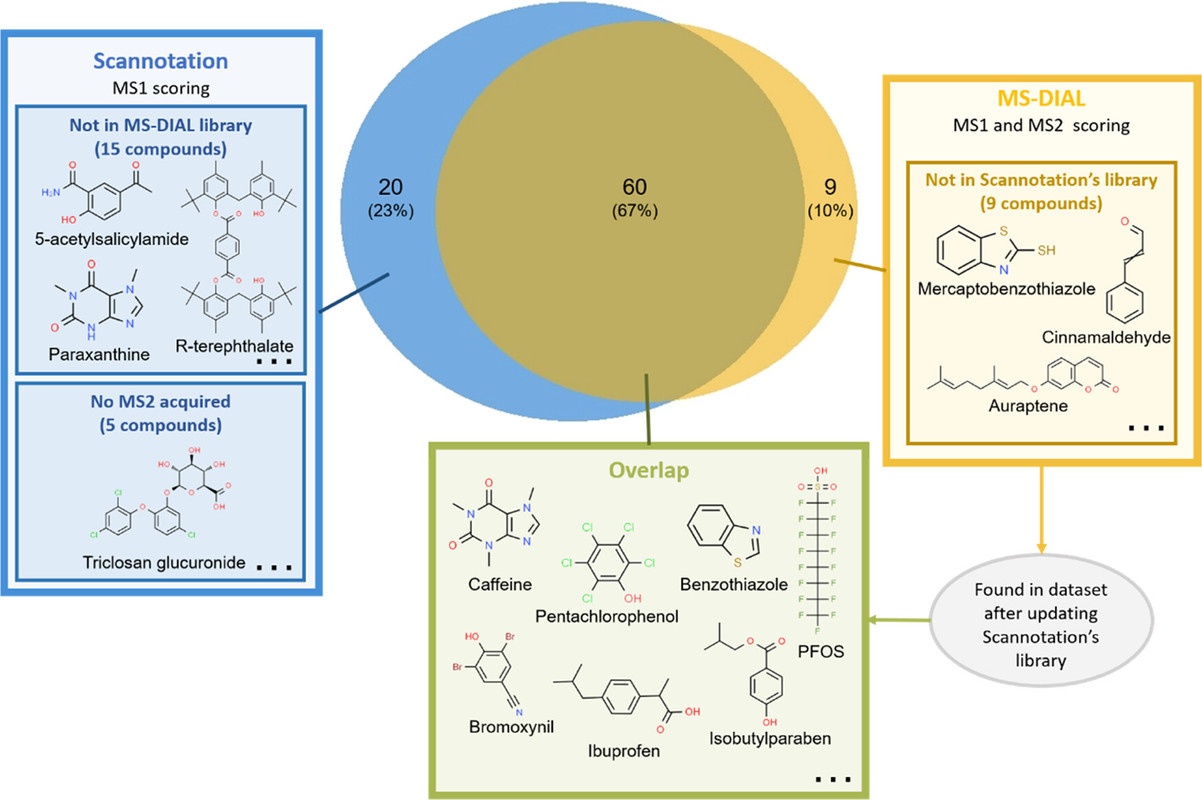
The caption:
The penultimate figure in the text gives an idea of the "exposome" found in human subjects:
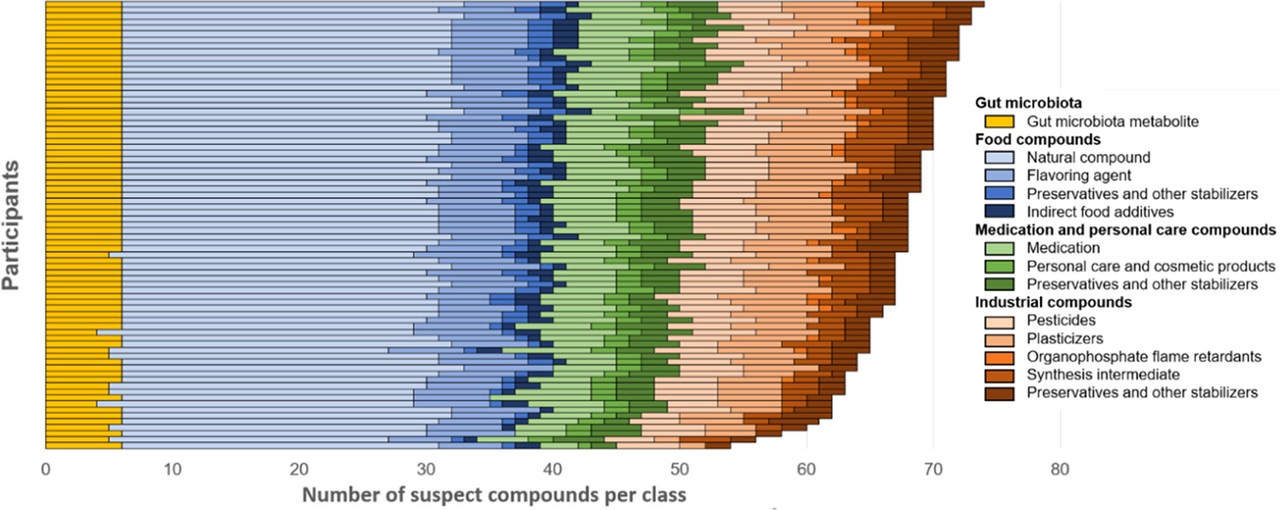
The caption:
An interesting, if disturbing, paper, I think.
Have a nice weekend.
moniss
(8,550 posts)data would look if we were able to do this on a person who lived in the early 1800's before all of the industrial exposures and personal care products. I know people who dump all kinds of stuff on themselves and their hair on a daily basis and then tell me how they have "healthy" skin and hair.